We have proposed a stochastic formulation enabling the representation of model-form uncertainties in MD simulations, given a set of model candidates identified through domain expertise or learned using machine learning (ML). The approach relies on a reduced-order model, the projection basis of which is randomized on a subset of the Stiefel manifold characterized by a set of linear constraints defining, e.g., Dirichlet boundary conditions in the physical space.
Figures below show an application to an argon crystal melt; see the figure below.
Image
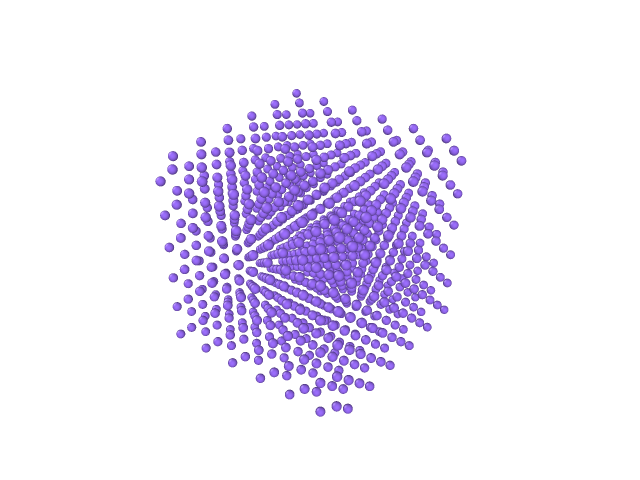
| Image
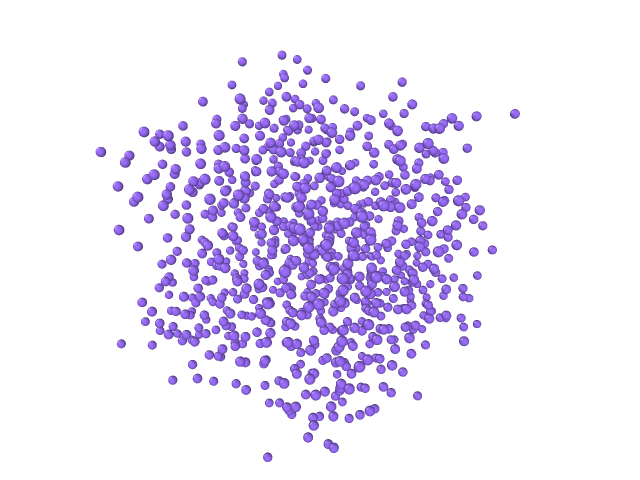
|
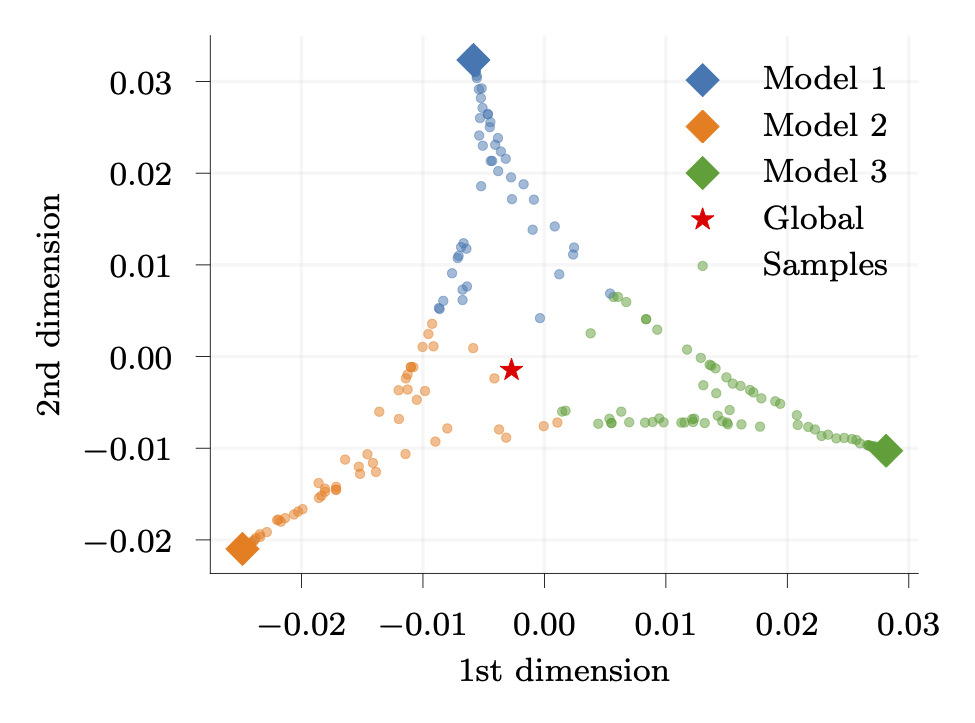
The approach allows for sampling over the matrix manifold, within the convex hull defined by the model candidates:
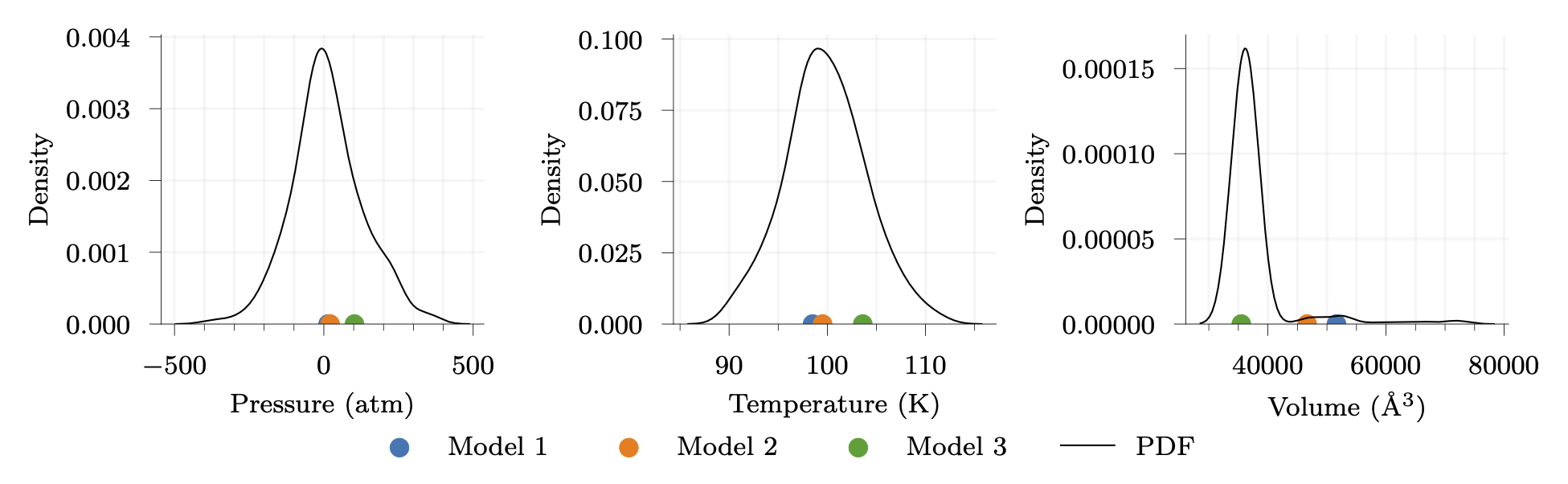
Uncertainty propagation is then conducted to quantify the impact on macroscopic properties:
Additional details can be found in the following papers:
- H. Zhang and J. Guilleminot, A Riemannian stochastic representation for quantifying model uncertainties in molecular dynamics simulations, Computer Methods in Applied Mechanics and Engineering, 403, 115702 (2023)
- S. Kounouho, R. Dingreville, and J. Guilleminot, Stochastic symplectic reduced-order modeling for model-form uncertainty quantification in molecular dynamics simulations in various statistical ensembles, Computer Methods in Applied Mechanics and Engineering, 431, 117323 (2024)